Monitoreo y Predicción de Enfermedades Infecciosas a través del Análisis de Redes Sociales
DOI:
https://doi.org/10.17488/RMIB.47.SI-TAIH.1529Palabras clave:
COVID-19, Redes sociales, BERT, Gompertz, Vigilancia epidemiológicaResumen
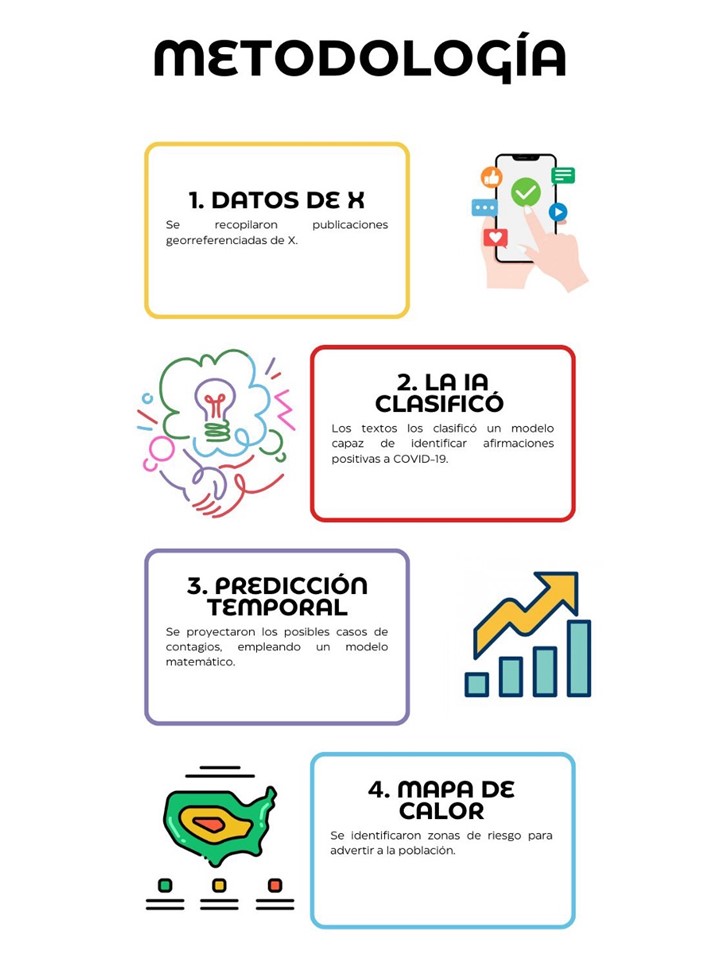
Este artículo presenta un enfoque integral para el monitoreo y la predicción de enfermedades infecciosas a partir de datos de redes sociales, centrándose principalmente en la pandemia de COVID-19. A través de la adquisición y el análisis de tweets georreferenciados se capturan señales tempranas de contagio, complementando de este modo los métodos tradicionales de vigilancia epidemiológica. El núcleo del sistema de clasificación se basa en el modelo BERT, el cual permite identificar automáticamente afirmaciones de infección, mientras que la función de Gompertz se utiliza para estimar el crecimiento de los contagios en horizontes de 1 a 5 días. La correlación significativa entre los datos extraídos de Twitter y las cifras oficiales reportadas por la Secretaría de Salud demuestra la solidez del método, además de evidenciar el potencial de los datos de las redes sociales para anticipar brotes y optimizar la toma de decisiones. Asimismo, la inclusión de datos geoespaciales posibilita la identificación de zonas con mayor nivel de riesgo, aportando un recurso valioso para las autoridades de salud y la población en general. Con ello se promueve una respuesta más rápida y coordinada ante futuras amenazas sanitarias.
Descargas
Citas
SAS, “Solutions for real-time disease surveillance and data-driven decision making.” Accessed: Oct. 31, 2024. [Online]. Available: https://www.sas.com/es_mx/insights/articles/analytics/situational-awareness-guides-our-responses---routine-to-crisis.html
Sasha Walek, “New COVID Local Risk Index Identifies Neighborhoods at Highest Risk of Pandemic’s Impact.” Accessed: Oct. 31, 2024. [Online]. Available: https://nyulangone.org/news/new-covid-local-risk-index-identifies-neighborhoods-highest-risk-pandemics-impact
S. Masri et al., “Use of Twitter data to improve Zika virus surveillance in the United States during the 2016 epidemic,” BMC Public Health, vol. 19, no. 1, Jun. 2019, doi: https://doi.org/10.1186/s12889-019-7103-8
J. O. Arjona, “Geolocated Social Networks And Covid-19: Analysis Of The Temporal And Spatial Activity Of Twitter Users In Spain During The Pandemic,” GeoFocus, vol. 2022, no. 30, pp. 25–47, Dec. 2022, doi: https://doi.org/10.21138/GF.789.
M. U. Hoque et al., “Analyzing Tweeting Patterns and Public Engagement on Twitter During the Recognition Period of the COVID-19 Pandemic: A Study of Two U.S. States,” IEEE Access, vol. 10, pp. 72879–72894, 2022, doi: https://doi.org/10.1109/ACCESS.2022.3189670.
J. S. P. Tulloch, R. Vivancos, R. M. Christley, A. D. Radford, and J. C. Warner, “Mapping tweets to a known disease epidemiology; a case study of Lyme disease in the United Kingdom and Republic of Ireland,” J Biomed Inform X, vol. 4, Dec. 2019, doi: https://doi.org/10.1016/j.yjbinx.2019.100060.
S. Chae, S. Kwon, and D. Lee, “Predicting infectious disease using deep learning and big data,” Int J Environ Res Public Health, vol. 15, no. 8, Aug. 2018, doi: https://doi.org/10.3390/ijerph15081596.
M. Birjali, A. Beni-Hssane, and M. Erritali, “Machine Learning and Semantic Sentiment Analysis based Algorithms for Suicide Sentiment Prediction in Social Networks,” in Procedia Computer Science, Elsevier B.V., 2017, pp. 65–72. doi: https://doi.org/10.1016/j.procs.2017.08.290.
D. Reynard and M. Shirgaokar, “Harnessing the power of machine learning: Can Twitter data be useful in guiding resource allocation decisions during a natural disaster?,” Transp Res D Transp Environ, vol. 77, pp. 449–463, Dec. 2019, doi: https://doi.org/10.1016/j.trd.2019.03.002.
Y. Gu, Y. Yao, W. Liu, and J. Song, “We know where you are: Home location identification in location-based social networks,” in 2016 25th International Conference on Computer Communications and Networks, ICCCN 2016, Institute of Electrical and Electronics Engineers Inc., Sep. 2016. doi: https://doi.org/10.1109/ICCCN.2016.7568598.
R. P. D. Redondo, C. Garcia-Rubio, A. F. Vilas, C. Campo, and A. Rodriguez-Carrion, “A hybrid analysis of LBSN data to early detect anomalies in crowd dynamics,” Future Generation Computer Systems, vol. 109, pp. 83–94, 2020, doi: https://doi.org/10.1016/j.future.2020.03.038.
O. Alonso, V. Kandylas, S. E. Tremblay, and S. Whiting, “Answering recreational web searches with relevant things to do results,” Inf Process Manag, vol. 57, no. 2, p. 102184, 2020, doi: https://doi.org/10.1016/j.ipm.2019.102184.
X. Xiong, S. Qiao, Y. Li, N. Han, G. Yuan, and Y. Zhang, “A point-of-interest suggestion algorithm in Multi-source geo-social networks,” Eng Appl Artif Intell, vol. 88, no. September 2019, p. 103374, 2020, doi: https://doi.org/10.1016/j.engappai.2019.103374.
W. Shi, D. Liu, J. Yang, J. Zhang, S. Wen, and J. Su, “Social bots’ sentiment engagement in health emergencies: A topic-based analysis of the covid-19 pandemic discussions on twitter,” Int J Environ Res Public Health, vol. 17, no. 22, pp. 1–19, Nov. 2020, doi: https://doi.org/10.3390/ijerph17228701.
V. Suarez-Lledo and J. Alvarez-Galvez, “Assessing the Role of Social Bots During the COVID-19 Pandemic: Infodemic, Disagreement, and Criticism,” J Med Internet Res, vol. 24, no. 8, Aug. 2022, doi: https://doi.org/10.2196/36085.
J. Jiang, X. Ren, and E. Ferrara, “Social Media Polarization and Echo Chambers in the Context of COVID-19: Case Study,” JMIRx Med, vol. 2, no. 3, p. e29570, Aug. 2021, doi: https://doi.org/10.2196/29570.
J. Xue et al., “Twitter discussions and emotions about the COVID-19 pandemic: Machine learning approach,” J Med Internet Res, vol. 22, no. 11, Nov. 2020, doi: https://doi.org/10.2196/20550.
A. Z. Klein, S. Kunatharaju, K. O’Connor, and G. Gonzalez-Hernandez, “Automatically Identifying Self-Reports of COVID-19 Diagnosis on Twitter: An Annotated Data Set, Deep Neural Network Classifiers, and a Large-Scale Cohort,” J Med Internet Res, vol. 25, 2023, doi: https://doi.org/10.2196/46484.
Sarker, S. Lakamana, W. Hogg-Bremer, A. Xie, M. A. Al-Garadi, and Y.-C. Yang, “Self-reported COVID-19 symptoms on Twitter: An analysis and a research resource,” Apr. 22, 2020. doi: https://doi.org/10.1101/2020.04.16.20067421.
E. Ferrara, “What types of COVID-19 conspiracies are populated by Twitter bots?,” First Monday, May 2020, doi: https://doi.org/10.5210/fm.v25i6.10633.
P. Castioni, G. Andrighetto, R. Gallotti, E. Polizzi, and M. De Domenico, “The voice of few, the opinions of many: Evidence of social biases in Twitter COVID-19 fake news sharing,” R Soc Open Sci, vol. 9, no. 10, Oct. 2022, doi: https://doi.org/10.1098/rsos.220716.
J. Mellon and C. Prosser, “Twitter and Facebook are not representative of the general population: Political attitudes and demographics of british social media users,” Research and Politics, vol. 4, no. 3, Jul. 2017, doi: https://doi.org/10.1177/2053168017720008.
S. Wojcik and A. Hughes, “Sizing Up Twitter Users For Media Or Other Inquiries,” 2019. [Online]. Available: www.pewresearch.org.
Harmon, “Tweepy,” Twitter for Python. Accessed: Apr. 08, 2025. [Online]. Available: https://www.tweepy.org
J. Devlin, M.-W. Chang, K. Lee, K. T. Google, and A. I. Language, “BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding.” [Online]. Available: https://github.com/tensorflow/tensor2tensor
A. Rogers, O. Kovaleva, and A. Rumshisky, “A Primer in BERTology: What We Know About How BERT Works”, doi: https://doi.org/10.1162/tacl_a_00349
K. Kaur and P. Kaur, “BERT-CNN: Improving BERT for Requirements Classification using CNN,” in Procedia Computer Science, Elsevier B.V., 2022, pp. 2604–2611. doi: https://doi.org/10.1016/j.procs.2023.01.234.
Pedro Wences, “Monitoreo epidemiológico de COVID-19 con datos de X,” CENIDET. Accessed: May 11, 2025. [Online]. Available: https://github.com/wopjk/MonitoreoCovidTwitterX
P. Winsor, “The Gompertz Curve As A Growth Curve,” 1932. [Online]. Available: https://doi.org/10.1073/pnas.18.1.1
K. M. C. Tjørve and E. Tjørve, “The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family,” PLoS One, vol. 12, no. 6, Jun. 2017, doi: https://doi.org/10.1371/journal.pone.0178691.
E. Pelinovsky et al., “Gompertz model in COVID-19 spreading simulation,” Chaos Solitons Fractals, vol. 154, Jan. 2022, doi: https://doi.org/10.1016/j.chaos.2021.111699.
CONAHCYT, “Covid-19 México,” CONAHCYT. Accessed: May 11, 2025. [Online]. Available: https://datos.covid-19.conacyt.mx
A. Tarafdar, J. Mahato, R. K. Upadhyay, and P. Bhattacharya, “Exploring the synergy of media awareness and quarantine classes in SiSAQEIH model for pandemic control: A Deep LSTM-RNN predictions,” Physica D, vol. 474, p. 134563, Apr. 2025, doi: https://doi.org/10.1016/j.physd.2025.134563.
W. Yang and X. Chang, “Time series analysis and prediction of the trends of COVID-19 epidemic in Singapore based on machine learning,” Computer Methods and Programs in Biomedicine Update, vol. 7, Jan. 2025, doi: https://doi.org/10.1016/j.cmpbup.2025.100190.
A. Chhabra et al., “Sustainable and intelligent time-series models for epidemic disease forecasting and analysis,” Sustainable Technology and Entrepreneurship, vol. 3, no. 2, May 2024, doi: https://doi.org/10.1016/j.stae.2023.100064.
S. Y. Ilu and R. Prasad, “Improved autoregressive integrated moving average model for COVID-19 prediction by using statistical significance and clustering techniques,” Heliyon, vol. 9, no. 2, Feb. 2023, doi: https://doi.org/10.1016/j.heliyon.2023.e13483.
W. Angulo, J. M. Ramírez, D. De Cecchis, J. Primera, H. Pacheco, and E. Rodríguez-Román, “A modified SEIR model to predict the behavior of the early stage in coronavirus and coronavirus-like outbreaks,” Sci Rep, vol. 11, no. 1, Dec. 2021, doi: https://doi.org/10.1038/s41598-021-95785-y.
R. Ramalingam, A. J. Gnanaprakasam, and S. Boulaaras, “Stability and control analysis of COVID-19 spread in India using SEIR model,” Sci Rep, vol. 15, no. 1, Dec. 2025, doi: https://doi.org/10.1038/s41598-025-93994-3.
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2026 Revista Mexicana de Ingenieria Biomedica

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial 4.0.
Una vez que el artículo es aceptado para su publicación en la RMIB, se les solicitará al autor principal o de correspondencia que revisen y firman las cartas de cesión de derechos correspondientes para llevar a cabo la autorización para la publicación del artículo. En dicho documento se autoriza a la RMIB a publicar, en cualquier medio sin limitaciones y sin ningún costo. Los autores pueden reutilizar partes del artículo en otros documentos y reproducir parte o la totalidad para su uso personal siempre que se haga referencia bibliográfica al RMIB. No obstante, todo tipo de publicación fuera de las publicaciones académicas del autor correspondiente o para otro tipo de trabajos derivados y publicados necesitaran de un permiso escrito de la RMIB.











